Table of Contents
ToggleWhat is Plasmid?
The term plasmid refers to a small, circular, extrachromosomal double-stranded DNA molecule that exists naturally in many bacteria, and in some archaea and eukaryotic organisms. Plasmids are physically distinct from chromosomal DNA but possess the ability to replicate independently due to their own origin of replication (Ori).
These mobile genetic elements often carry genes that provide selective advantages, such as antibiotic resistance, virulence, or metabolic capabilities, enabling bacteria to adapt and survive under diverse environmental conditions.
Depending on the bacterial species, plasmid sizes can vary from a few hundred base pairs to several hundred kilobases. During cell division, plasmids are replicated and distributed to daughter cells. Some plasmids can also be transferred between bacteria through a process known as bacterial conjugation, facilitating horizontal gene transfer and genetic diversity.
History of Plasmids
Early plasmid research in Escherichia coli and Staphylococcus aureus introduced the concept of episomes — genetic elements capable of existing freely or integrating into chromosomes.
In the 1950s and 1960s, scientists discovered R (resistance) plasmids, which carried multiple antibiotic resistance genes and could spread via conjugation. This led to extensive studies that revealed plasmids often contain two main components:
- Resistance Transfer Factor (RTF) – Controls replication and transfer.
- Resistance Determinants (r) – Encode antibiotic resistance.
During the 1970s, plasmids were classified by incompatibility grouping, emphasizing differences in replication control and partitioning mechanisms. Research expanded to include Col, Ti, degradative, and virulence plasmids, enhancing our understanding of replication, transfer, and evolution.
Today, plasmids play key roles in medicine, agriculture, biotechnology, and environmental microbiology.
Properties of Plasmids
1. Replication Mechanism
- The majority of plasmids replicate similarly to bacterial chromosomes.
- The DNA opens at their replication origin, initiating bidirectional replication.
- The circular plasmid’s two replication forks move in opposing directions until they meet.
- The other replication mechanism is the rolling circle replication mechanism. It starts by nipping the replication origin and unwinding one strand, using the other, still circular, strand as a template for DNA production.
- Some plasmids, such the F plasmid found in E. coli, have the ability to travel between microorganisms. These plasmids have two different replication origins.
2. Copy Number
- After division, the number of plasmid copies in a bacterial cell is referred to as the copy number.
- High copy plasmids (10–100 copies) are less restricted than low copy plasmids (1–2 copies).
- Particular genes regulate plasmid replication via three primary mechanisms:
- Antisense RNA
- Replication protein binding to iterons
- A combination of antisense RNA and protein control.
3. Incompatibility
- Closely related plasmids cannot coexist stably in the same host cell — a property known as plasmid incompatibility. Over time, one plasmid will dominate while the other is lost.
Structure of Plasmid
Plasmids are generally tiny (1–400 kb), circular, double-stranded DNA molecules that replicate outside of the bacterial chromosome. In some yeast and bacteria, plasmids may be linear, though the majority of them are spherical.

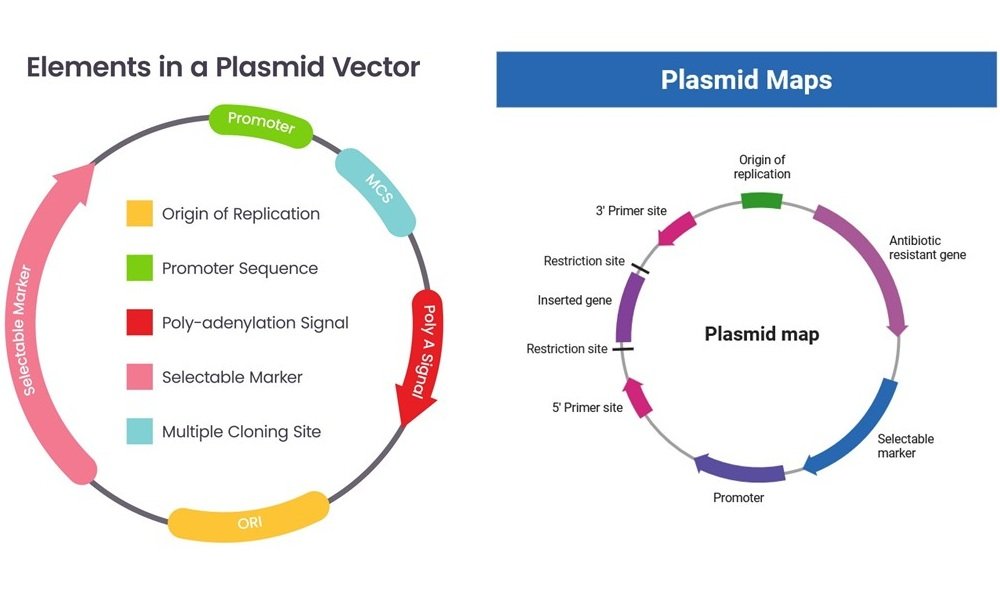
1. Origin of Replication (Ori)
- The precise sequence of nucleotides that starts replication.
- Has binding sites for replication proteins (Rep proteins).
- Identifies the copy number and compatibility group.
- It might be regulated by regulatory proteins, iterons, or antisense RNA.
2. Selectable Marker Genes
- Antibiotic resistance is often encoded (e.g., bla, Tet, Kan).
- Under certain circumstances, permit the selection of plasmid-containing cells.
3. Replication and Maintenance Genes
- Rep genes regulate replication initiation.
- Partitioning genes (parA, parB, parS) ensure even plasmid distribution during cell division.
- Toxin-antitoxin systems prevent plasmid loss from the population.
4. Accessory Genes
- Encode adaptive traits like:
- Virulence factors (Yersinia pYV plasmid).
- Metabolic pathways (hydrocarbon degradation).
- Symbiotic nitrogen fixation (Rhizobium plasmids).
5. Multiple Cloning Site (MCS)
- A synthetic DNA region with various restriction enzyme sites used for inserting foreign DNA during gene cloning.
6. Regulatory Elements
- Include promoters, operators, and terminators controlling gene expression.
- May be inducible (e.g., lac promoter) or constitutive.
7. Mobility and Conjugation Genes
- tra genes encode proteins required for pilus formation and DNA transfer.
- mob genes assist in mobilization when a conjugative plasmid is present.
8. Structural Variations
- Conjugative plasmids: contain transfer genes.
- Non-conjugative plasmids: depend on other plasmids for transfer.
- Cryptic plasmids: possess replication genes but no known phenotypic traits.
Types of Plasmids
1. Fertility (F) Plasmids
- Contains transfer (tra) genes that facilitate gene exchange between bacteria through conjugation.
- Belong the large class of conjugative plasmids.
- Are episomes— plasmids that can integrate into chromosomal DNA.
- F+ bacteria: contain the F-plasmid.
- F– bacteria: lack the F-plasmid.
- Pair of F+ bacteria is produced when an F+ and F– bacterium conjugate.
- An F plasmid can only be carried by one bacterium at a time.
2. Resistance (R) Plasmids
- Have genes that aid bacteria in resisting antibiotics or poisons.
- Through conjugation, certain R plasmids are able to transmit themselves.
- Promote the transmission of antibiotic resistance across bacterial communities.
- For example: Cephalosporins are advised as an alternative to quinolones because Neisseria gonorrhoeae has become highly resistant to them.
- The proliferation of drug-resistant strains is accelerated by the overuse of antibiotics, for example, for UTIs.
3. Col Plasmids (Colicin plasmids)
- Carry genes for bacteriocins (like colicins), which are proteins that destroy other bacteria.
- By removing competitors, give the host bacteria a competitive advantage.
- For example: the ColE1 plasmid in E. coli is responsible for the synthesis of bacteriocins.
4. Virulence Plasmids
- Change harmless bacteria into harmful ones.
- Have genes that allow the bacteria to induce illness.
- For instance, some strains of E. coli that have virulence plasmids can cause severe diarrhea and vomiting.
- Additionally, Salmonella enterica has virulence plasmids.
5. Degradative Plasmids
- Facilitate the breakdown of unusual organic substances like salicylic acid, toluene, xylene, and camphor.
- Have genes for unique enzymes that break down these substances.
- They are able to transfer to other bacteria since they are conjugative.
Replication Mechanisms of Plasmids
Plasmids replicate autonomously due to their own Ori sequence.
Enzymatic Involvement:
- Small plasmids→ Rely entirely on host cell enzymes.
- Large plasmids → May carry genes coding for plasmid-specific replication enzymes.
Gene composition:
- Typically, there are less than 30 genes.
- The beginning of replication is controlled.
- Sharing plasmids between progeny cells.
- Plasmid genes are typically not necessary for the host’s survival; in fact, plasmid-free bacteria frequently behave normally.
Replication Speed:
- Replication is quick because of the small size; it could only take 1/10 or less of the cell division cycle.
Main Replication Mechanisms
1. Theta (Θ) Replication
Found in Gram-Negative bacteria.
Procedure:
- Beginning at the origin of replication (ori).
- Replication forks moving in bidirectional.
- The Greek letter theta (Θ) is similar to the intermediate.
- Variation: Unidirectional theta replication is used by some Gram-negative plasmids.
- For instance, the ColE1 plasmid.
2. Rolling Circle (RC) Replication
Found often in the majority of plasmids in Gram-positive bacteria. Similar to: The replication of bacteriophage φX174.
Procedure:
- By nicking a single strand, 3′OH is made available for DNA synthesis.
- The previous strand is replaced by the synthesis of a new strand.
- The template for complementary strand synthesis is the displaced strand.
- Makes several copies quickly.
3. Strand Displacement Replication
Plasmids having a wide host range (e. g., the IncQ family).
Procedure:
- One strand is displaced by DNA synthesis without the formation of a theta intermediate.
- Replication is possible without double-strand breaks due to continuous displacement.
Replication in Linear Plasmids
Present in both Gram-negative and Gram-positive bacteria.
Two structural forms:
- Hairpin endings at each end point → replicate via concatemeric intermediates.
- Protein-bound 5′ ends → replicate via protein-priming mechanism (similar to bacteriophage φ29).
A unique situation exists when certain linear plasmids, even those with terminal proteins, begin replicating from an internal origin.
Applications of Plasmids
Plasmids have become indispensable tools in biotechnology, medicine, and genetic engineering.
- Gene Cloning: Gene cloning Plasmids are used as vectors to introduce and replicate particular DNA sequences.
- Protein synthesis: Used to produce proteins like insulin, growth hormones, and enzymes in host cells.
- The creation of vaccines: DNA vaccines trigger an immune response by using plasmids that contain antigen genes.
- Gene therapy: Deliver therapeutic genes into human cells to correct genetic defects.
- Metabolic engineering: Change bacterial metabolic pathways to produce biofuels, bioplastics, or drugs.
- Gene function research: Use reporter genes like GFP to monitor expression and control.
- Agricultural biotechnology: Introducing advantageous genes into plants in order to increase production or boost pest resistance.
- Bioremediation: Bioremediation involves inserting genes that degrade environmental contaminants into bacteria.
- Antibiotic Resistance Studies: Studies of antibiotic resistance, which are utilized in research to comprehend and track the movement of resistance genes.
- Synthetic biology: Serve as modular genetic components for creating new biological systems.
Conclusion
Research on F, R, and Col plasmids in E. coli and Staphylococcus aureus established the foundation for modern plasmid biology. The discovery of R plasmids in the mid-20th century revolutionized our understanding of gene transfer and antibiotic resistance.
Subsequent classification and molecular studies revealed diverse plasmid types—toxin, degradative, Ti, and Rhizobium plasmids—each vital to bacterial adaptability.
Today, plasmids are crucial in genetic engineering, biotechnology, medicine, and environmental science, providing essential tools for innovation and sustainable development.
Frequently Asked Questions (FAQs)
Q1. What is a plasmid in microbiology?
A plasmid is a small, circular DNA molecule that replicates independently of chromosomal DNA and often carries genes beneficial to bacterial survival.
Q2. What is the main function of plasmids?
Plasmids provide bacteria with adaptive traits such as antibiotic resistance, virulence, metabolic pathways, and toxin production.
Q3. How are plasmids used in genetic engineering?
Plasmids act as cloning vectors that carry foreign genes into host cells for gene expression, protein production, or gene therapy.
Q4. What are the main types of plasmids?
The five major types include F (fertility), R (resistance), Col (bacteriocin), Virulence, and Degradative plasmids.
Q5. What is the difference between chromosomal DNA and plasmid DNA?
Chromosomal DNA contains essential genes for cell survival, while plasmid DNA carries non-essential genes that provide selective advantages.
Q6. Why are plasmids important in biotechnology?
Plasmids are vital tools for recombinant DNA technology, vaccine development, gene therapy, and bioremediation.
Also Read
- Microbiology of Fermented Foods
- How Microbiology Helps in Vaccine Development: A Comprehensive Guide
- Branches of Microbiology: An Overview of Key Fields
- Importance of Microbiology: Applications, Scope, and Benefits
- Microbiology Experiments for Students: A Complete Guide
- Parasitology: An Overview of Parasites, Diseases, and Host Interactions
- Microbiology: From Microorganisms to Career Opportunities
- Molecular Blotting: Techniques, Uses, and Significance
- Ribonucleic acid (RNA): Types, Structure & their Function
- Antimicrobial Resistance (AMR) Mechanisms and Alternatives
- Spirulina: The Superfood Microalga with Limitless Potential
- Microbiology Notes
- Bacteriology Quiz

