Table of Contents
ToggleIntroduction to Antimicrobial Resistance (AMR)
Antimicrobial Resistance (AMR) occurs when microorganisms—such as bacteria, viruses, fungi, and parasites—evolve to survive exposure to drugs that once killed or inhibited them. It poses a major threat to global health, food security, and development. According to WHO, by 2050, AMR could cause 10 million deaths annually if not controlled.
The rise of antibiotic-resistant pathogens like MRSA, CRE, and XDR-TB is a global health crisis.
Current Focus Areas in AMR Research:
- Understanding genetic mutations and plasmid-mediated resistance.
- CRISPR-based approaches to disrupt resistance genes.
- Phage therapy and antimicrobial peptides as alternative treatments.
- Surveillance systems using whole-genome sequencing (WGS) for AMR tracking.
Mechanisms of Antimicrobial Resistance
Antimicrobial resistance arises when microorganisms evolve strategies to neutralize, evade, or resist the effects of drugs designed to kill or inhibit them. These mechanisms can be innate (natural) or acquired through genetic changes or horizontal gene transfer.
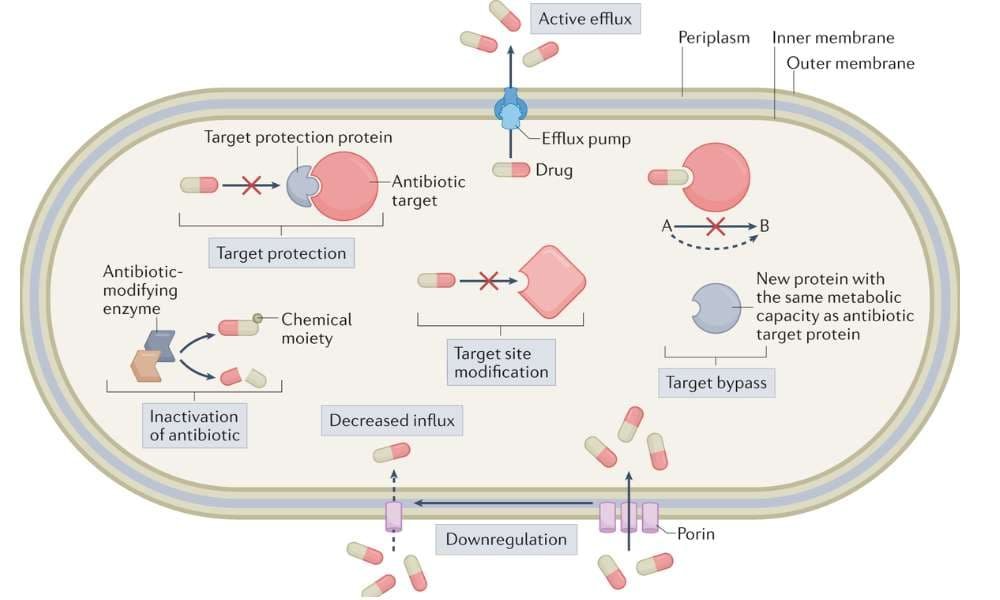
Microorganisms use several strategies to evade antimicrobial drugs
- Enzymatic Degradation or Modification of the Drug
- Target Site Alteration
- Efflux Pumps
- Reduced Membrane Permeability
- Biofilm Formation
- Bypass of Metabolic Pathways
- Horizontal Gene Transfer (HGT)
Enzymatic Degradation or Modification of the Drug
Microorganisms produce enzymes that break down or inactivate antibiotics before they can act.
Example:
- β-lactamases cleave the β-lactam ring of penicillins and cephalosporins.
- Carbapenemases (e.g., KPC, NDM-1) degrade even the most potent β-lactams.
- Aminoglycoside-modifying enzymes add chemical groups (e.g., acetylation), rendering the drug ineffective.
Target Site Alteration
Mutations or gene acquisitions can lead to changes in the drug’s binding site, reducing the drug’s ability to bind and inhibit the target.
Examples:
- Methicillin-resistant Staphylococcus aureus (MRSA) produces an altered penicillin-binding protein (PBP2a) with low affinity for β-lactams.
- Macrolide resistance through methylation of 23S rRNA in the 50S ribosome subunit.
- Fluoroquinolone resistance via mutations in DNA gyrase and topoisomerase IV.
Efflux Pumps
Efflux pumps are membrane proteins that actively expel antibiotics from the microbial cell, preventing accumulation to toxic levels.
Examples:
- AcrAB-TolC system in E. coli for multidrug resistance.
- MexAB-OprM efflux system in Pseudomonas aeruginosa.
- These pumps can confer resistance to tetracyclines, fluoroquinolones, macrolides, and chloramphenicol.
Reduced Membrane Permeability
Some bacteria, especially Gram-negative species, reduce the permeability of their outer membrane to limit antibiotic entry.
Examples:
- Porin loss or mutation in Klebsiella pneumoniae reduces uptake of β-lactams.
- Decreased permeability combined with efflux pumps often results in high-level resistance.
Biofilm Formation
Microorganisms in biofilms are 100–1000 times more resistant to antibiotics than planktonic (free-floating) cells.
How biofilms confer resistance:
- Physical barrier: biofilm matrix slows antibiotic penetration.
- Altered microenvironment: pH and oxygen gradients reduce drug efficacy.
- Dormant cells: persister cells are metabolically inactive and unaffected by antibiotics targeting growth.
Bypass of Metabolic Pathways
Bacteria can develop alternative metabolic pathways to avoid inhibition by specific drugs.
Example:
- Resistance to sulfonamides occurs when bacteria acquire alternative dihydropteroate synthase enzymes that bypass folic acid synthesis inhibition.
Horizontal Gene Transfer (HGT)
Resistance genes can be acquired from other organisms via:
A. Conjugation
- Direct transfer of plasmids carrying resistance genes between bacteria through a pilus.
- Example: R plasmids carrying multiple resistance genes.
B. Transformation
- Uptake of free DNA fragments from the environment.
- Common in Streptococcus and Neisseria species.
C. Transduction
- Bacteriophages transfer resistance genes between bacterial cells.
- Plays a role in spreading toxin and resistance genes in clinical pathogens.
Summary Table: AMR Mechanisms
| Mechanism | Example Organisms | Antibiotics Affected |
| Enzymatic degradation | E. coli, K. pneumoniae, P. aeruginosa | β-lactams, aminoglycosides |
| Target modification | S. aureus, M. tuberculosis | β-lactams, macrolides, fluoroquinolones |
| Efflux pumps | E. coli, P. aeruginosa, S. aureus | Multiple classes |
| Reduced permeability | K. pneumoniae, N. gonorrhoeae | β-lactams, quinolones |
| Biofilm formation | P. aeruginosa, S. epidermidis | Most antibiotics |
| Bypass pathways | E. coli, Shigella spp. | Sulfonamides, trimethoprim |
| Horizontal gene transfer | Many clinical isolates | Multidrug resistance |
Superbugs and WHO Priority Pathogens
The WHO has published a list of priority pathogens resistant to multiple antibiotics, including:
- Carbapenem-resistant Acinetobacter baumannii.
- Multidrug-resistant Pseudomonas aeruginosa.
- Vancomycin-resistant Enterococci (VRE).
- Extensively Drug-Resistant Tuberculosis (XDR-TB).
Alternative Strategies to Combat AMR
With the development of new antibiotics slowing and multidrug-resistant “superbugs” on the rise, scientists are urgently exploring novel, non-traditional strategies to overcome AMR. These approaches target bacterial survival and resistance mechanisms using biological, technological, and ecological methods.
Phage Therapy
What it is:
The use of bacteriophages—viruses that infect and lyse specific bacteria—as an alternative to antibiotics.
Why it matters:
- Phages are highly specific, killing only targeted bacterial strains.
- Can be customized into personalized phage cocktails for chronic infections like cystic fibrosis, diabetic wounds, or UTIs.
- Useful against biofilm-associated infections.
Challenges:
- Regulatory barriers and phage resistance development.
- Need for precise bacterial identification before therapy.
Antimicrobial Peptides (AMPs)
What they are:
Short, naturally occurring peptides from humans, plants, and animals that disrupt microbial membranes.
Key features:
- Broad-spectrum activity.
- Low tendency to induce resistance.
- Can act against both planktonic cells and biofilms.
Examples:
- Defensins (human), magainins (frog skin), and cathelicidins.
Nano-Antibiotics
What they are:
Nanoparticles engineered to improve antibiotic delivery, reduce toxicity, or enhance antimicrobial effects.
Types of nanoparticles in use:
- Silver and zinc oxide: inherently antimicrobial.
- Lipid-based nanoparticles: improve drug solubility and targeting.
- Magnetic nanoparticles: used for targeted therapy and diagnostics.
Advantages:
- Bypass resistance mechanisms.
- Penetrate biofilms and infected tissues effectively.
Quorum Sensing Inhibitors (QSIs)
What it is:
Compounds that disrupt bacterial communication (quorum sensing), which regulates virulence, toxin production, and biofilm formation.
Benefits:
- Reduce bacterial pathogenicity without killing them (lower resistance pressure).
- Can enhance antibiotic efficacy when used in combination therapy.
Examples:
- Halogenated furanones, ajoene (from garlic), and synthetic analogs.
Probiotics and Microbiome Modulation
Concept:
Using beneficial microbes to outcompete or suppress resistant pathogens, particularly in the gut, skin, and oral microbiota.
Strategies include:
- Fecal Microbiota Transplantation (FMT): Restores healthy gut flora.
- Use of targeted probiotics or synbiotics.
- Enhancing colonization resistance post-antibiotic treatment.
Applications:
- Prevention of recurrent Clostridioides difficile infections.
- Modulating resistance gene carriage in the gut.
Repurposing Existing Drugs
What it means:
Using non-antibiotic drugs that show antimicrobial properties in new contexts.
Examples:
- Disulfiram (used for alcohol addiction) inhibits Borrelia burgdorferi.
- Chloroquine, metformin, and statins show immunomodulatory or antimicrobial activity in certain infections.
Advantages:
- Reduced development costs.
- Faster regulatory approval.
Recent Research & Innovations (2024–2025)
New technologies are driving innovation in the AMR space:
AI & Deep Learning Models
- Used to predict novel antibiotic structures.
- Identify resistance genes in large-scale metagenomic data.
Rapid Point-of-Care Diagnostics
- CRISPR-based tests (e.g., SHERLOCK, DETECTR) for resistance gene detection.
- MALDI-TOF mass spectrometry for rapid bacterial identification and resistance profiling.
Synthetic Biology & Engineered Bacteria
- Designing smart probiotics that secrete antimicrobial substances.
- Genetically engineered bacteria to outcompete resistant strains in the microbiome.
Global Action Plans on Antimicrobial Resistance
Organizations like WHO, CDC, and UNEP emphasize the One Health Approach, connecting human, animal, and environmental health:
- Reducing antibiotic use in agriculture.
- Promoting antibiotic stewardship in hospitals.
- Encouraging surveillance and global collaboration.
Conclusion
AMR is one of the most urgent microbiological challenges of our time. While resistance mechanisms continue to evolve, research into innovative alternatives—from phage therapy to nanotechnology—offers hope. Multidisciplinary efforts, combining genomics, AI, clinical practice, and policy-making, are crucial to tackling this silent pandemic.
Also Read
Reference and Sources
- https://pmc.ncbi.nlm.nih.gov/articles/PMC10340576/
- https://www.sciencedirect.com/science/article/pii/S0169409X05000980
- https://www.who.int/news/item/29-04-2019-new-report-calls-for-urgent-action-to-avertantimicrobial-resistance-crisis
- https://pubmed.ncbi.nlm.nih.gov/31687804/
- https://www.researchgate.net/figure/Permeability-of-E-coli-P-aeruginosa-S-aureus-and-Lmonocytogenes-to-propidium_fig3_235933847

